- 1Rice Research Institute, Yunnan Agricultural University, Kunming, China
- 2Department of Chemistry, University of Science and Technology, Bannu, Pakistan
- 3State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, Yunnan Agricultural University, Kunming, China
- 4Key Laboratory for Agricultural Biodiversity and Pest Management of China Education Ministry, Yunnan Agricultural University, Kunming, China
The embryo sac plays a vital role in sexual reproduction of angiosperms. LysM domain containing proteins with multiple lysin motifs are widespread proteins and are involved in plant defense responses against fungal chitins and bacterial peptidoglycans. Various studies have reported the role of LysM domain-containing proteins in plant defense mechanisms but their involvement in sexual reproduction remains largely unknown. Here, we report the involvement of a LysM domain-containing gene, EMBRYO SAC 1 (OsEMSA1), in the sexual reproduction of rice. The gene encoded a LysM domain-containing protein that was necessary for embryo sac development and function. The gene was expressed in root, stem, leaf tissues, panicle and ovaries and had some putative role in hormone regulation. Suppression of OsEMSA1 expression resulted in a defective embryo sac with poor differentiation of gametophytic cells, which consequently failed to attract pollen tubes and so reduced the panicle seed-setting rate. Our data offers new insight into the functions of LysM domain-containing proteins in rice.
Introduction
Sexual reproduction in plants begins with gametogenesis. Female gametogenesis is a biphasic process involving complex, tightly orchestrated developmental mechanisms, sequential cell divisions, subsequent nuclei migration, cellularization, and programmed cell death leading to the development of the megagametophyte known as the embryo sac (Reiser and Fischer, 1993; Drews et al., 1998). Female gametophyte development and function depends on the activities of many genes expressed either within itself or in the surrounding cells (Drews et al., 1998; Pagnussat et al., 2005). The female gametophyte, or embryo sac, develops coordinately with the sporophytic tissues of the ovule, making it an ideal model for the study of fundamental processes crucial to development (Drews et al., 1998). The embryo sac is considered pivotal in ensuring successful fertilization, embryogenesis and subsequent seed development (Pagnussat et al., 2005). In several previous studies, the role of phytohormones in controlling the female gametophyte has been well established (Deng et al., 2010; Bencivenga et al., 2012; Cheng et al., 2013). It was reported that manipulation of concentration of the phytohormone auxin results in defects of female gametophyte development (Pagnussat et al., 2009). Not only localized auxin biosynthesis but also auxin import are required for cellularization during embryo sac development (Panoli et al., 2015). Cytokinin was found to be indispensable for the male and female gamete development in Arabidopsis (Hirano et al., 2008; Kinoshita-Tsujimura and Kakimoto, 2011; Yamaki et al., 2011; Cheng et al., 2013). The regulatory interaction between auxin and cytokinin has also been linked to the development of the female gametophyte (Schaller et al., 2015).
The ability of the embryo sac to attract a pollen tube is crucial for effective fertilization. During fertilization, the pollen tube grows directionally inside the pistil and delivers the sperm to the embryo sac. The female sporophytic tissues facilitates the pollen tube's journey by providing multiple, stage specific, guidance signals along the pollen tube path (Wang et al., 1993; Cheung et al., 1995; Hulskamp et al., 1995; Ray et al., 1997; Fiebig et al., 2000; Mollet et al., 2000; Wu et al., 2000; Palanivelu et al., 2003; Palanivelu and Tsukamoto, 2012). The final phases of pollen tube guidance are controlled by the embryo sac (Higashiyama et al., 1998). The two synergid cells located in the embryo sac are supposed to secrete some chemotropic substances that attracts the pollen tube (Higashiyama et al., 2001). Recently, it was identified that several cell-surface receptors located on the pollen tube mediates the male perception of female attractant in Arabidopsis (Takeuchi and Higashiyama, 2016; Wang et al., 2016). The chemotropic substances secreted by synergids have been identified to be defensin-like LURE peptides (Okuda et al., 2009). Despite the importance of male-female communication in fertilization and subsequently seed and/or fruit production, this phenomenon is poorly understood in rice. As one of the most important crops and a model monocotyledonous plant, various stigma, pistil and pollen-specific, genes have been identified in the rice genome (Yoshida et al., 2005; Park et al., 2006; Li et al., 2007).
LysM are a family of carbohydrate-binding modules with multiple lysin motifs and are widely present in microbes, plants and animals (Shi et al., 2013). The multiple LysMs present in LysM-domain are separated from each other by some short spacer sequences (Buist et al., 2008). LysMs can be found at the end terminals or in the center of a protein (Buist et al., 2008). LysM containing proteins can be secreted proteins, membrane proteins, outer-membrane proteins, lipoproteins or cell wall bound proteins. LysM containing proteins bind to N-acetylglucosamine-containing carbohydrates, such as chitin, chitio-oligosaccharides and peptidoglycanwith various specificities (Akcapinar et al., 2015). Most of the LysM domain containing plant proteins belong to the class of RLKs (receptor like kinases) in plants (Wan, 2015). RLKs have a major role in cellular signaling in plants and most of the LysM domain-containing RLKs are involved in plant-microbial interactions leading to pathogen defense, symbiosis or suppression of host defense (Gust et al., 2012; Tanaka et al., 2013; Wan, 2015). Rice LysM protein CEBiP (chitin elicitor-binding protein) was shown to be involved in chitin recognition and activation of plant innate immunity against chitin (Kaku et al., 2006). The CEBiP protein has an extracellular domain that contains two LysMs. Another protein, OsCERK1 (chitin elicitor receptor kinase 1) function together with CEBiP and is involved in chitin triggered immunity in rice (Shimizu et al., 2010). Two dual acting lysM proteins, OsLYP4 and OsLYP6, were also found to be involved in perception and recognition of bacterial peptidoglycan and fungal chitin (Liu et al., 2012). Silencing of CEBiP, OsCERK1, LYP4, and LYP6 substantially increase susceptibility of O.sativa to microbial pathogenesis (Kaku et al., 2006; Shimizu et al., 2010; Liu et al., 2012). The LysM-containing receptor-like kinase1/chitin elicitor receptor kinase1 (LYK1/CERK1) of Arabidopsis was shown to be essential for chitin recognition (Miya et al., 2007; Wan et al., 2008; Willmann et al., 2011). Various studies have mentioned the role of LysM domain-containing proteins in plant innate immunity but there has been no report of their role on plant sexual reproduction.
Our previous study identified a rice Bsister-MADS Box gene, FEMALE-STERILE (FST), which is expressed in the sporophytic tissues of ovules and plays vital roles during ovule and early seed development (Lee et al., 2013). Based on the microarray data of the fst mutant, we selected the candidate gene OsEMSA1 (BankIt1920511, KX503265), a LysM domain-containing gene, located on chromosome 10, whose expression level was significantly down regulated at meiotic stage in rice panicles. This study aimed to explore the role and function of OsEMSA1 during sexual reproduction in rice. Our results showed that OsEMSA1 encoded a LysM domain-containing protein that was crucial for embryo sac development in rice. Our findings revealed an important role for a LysM domain-containing protein during sexual reproduction in rice.
Materials and Methods
Plant Material and Growth Condition
The wild type rice (O. sativa ssp. japonica cv. Ilmibyeo) was used for rice transformation in this study. Rice seeds were germinated in distilled water and grown in a greenhouse experiment field under natural growing conditions.
Gene Cloning, Characterization, and Bioinformatic Analysis
A 952-bp OsEMSA1 cDNA fragment was amplified from KOME clone by specific primers OsEMSA1-1F and OsEMSA1-1R (Supplementary Table 2). About 2.2-kb putative promoter upstream of the OsEMSA1 coding region fragment was amplified by PCR with primers OsEMSA1P-F and OsEMSA1P-R using Ilmibyeo genomic DNA as a template (Supplementary Table 2). Protein sequence and homology analysis was performed using NCBI databases. Protein sequence motifs were identified using the SMART program. Sequence alignments and the phylogenetic tree were constructed using MEGA6 and the neighbor-joining method.
Binary Vector Constructs and Transgenic Plant Development
To investigate the expression pattern of OsEMSA1, its promoter fragment was fused to the GUS reporter gene and subcloned into the binary vector DTV1 (the modified pCAMBIA1305.2 without enhancer) to yield the pOsEMSA1::GUS construct. To construct the OsEMSA1 RNAi vector (pCaMV35S×2::OsEMSA1-RNAi), a 117-bp intron fragment was used as a linker between a 170-bp gene-specific fragment (a 422-bp gene-specific fragment was generated synchronously) in the antisense and sense orientations. These reconstructed fragments were inserted into the DTV6 binary vector containing a double 35S promoter.
To investigate whether Os.51835 and Os.43929 are involved in hormone regulation, Os.51835 and Os.43929 RNAi binary vectors (pCaMV35S×2::Os51835-RNAi and pCaMV35S×2::Os43929-RNAi) were constructed, respectively.
All the constructs were introduced into Agrobacterium tumefaciens strain EHA105 and subsequently introduced into Ilmibyeo rice embryonic callus. Various T1 transgenic plants were generated: 4 lines (each line 10 plants) of pOsEMSA1::GUS transgenic plants, 8 lines (each line 10 plants) pCaMV35S×2::OsEMSA1-RNAi1 transgenic plants, 7 lines (each line 10 plants) pCaMV35S×2::OsEMSA1-RNAi2 transgenic plants, 6 lines (each line 10 plants) pCaMV35S×2::Os51835-RNAi T1 transgenic plants and 8 lines (each line 10 plants) pCaMV35S×2::Os43929-RNAi T1 transgenic plants were generated.
Pollen Viability and Germination Assay
To study pollen viability, spikelets were fixed in Carnoy's fixative solution (99% ethanol: chloroform: glacial acetic acid of 6:3:1) and stained in I2-KI and simplified Alexander's staining solution as mentioned previously (Peterson et al., 2010; Wang et al., 2012). Pollen grains from anthers were placed in 1% I2-KI staining solution and 1% simplified Alexander's staining solution for 20 min at room temperature to stain the pollen. Pollen grains that were round and stained black by I2-KI solution were considered fertile. Pollen grains that were stained purple or red by simplified Alexander's staining solution were considered viable.
Pollen germination and pollen tube growth were examined using aniline blue staining. Spikelets were collected during 30–60 min after flowering and then immediately placed in fixative solution (99% ethanol: glacial acetic acid of 3:1). The fixed sample was hydrated by passing through an ethanol series (70, 50, and 30% and distilled water) for a duration of 10 min for each step at room temperature. Pistils were excised, softened with 1 M NaOH at 60°C for 1 h and subsequently rinsed twice with distilled water, each for 10 min. Pollen tubes were stained with 0.1% (w/v) aniline blue in 100 mM K3PO4 buffer (pH 11) for 10 min in darkness. Samples were then visualized by UV microscopy (Mori et al., 2006).
Endogenous Hormone Assay
Endogenous contents of GA3 and IAA were determined using an ELISA hormone assay kit. The fresh tissues including root, stem, leaf and panicle were sampled at flowering stage and finely homogenized in 0.01 M PBS buffer (pH 7.4). The homogenate was centrifuged at 3,000 rpm for 5 min and the supernatant collected and preserved at −70°C. ELISA was performed as described in the protocol provided by the supplier (Plant Hormone Elisa Kit, Colorful Gene Biotechnology Co. Ltd., Wuhan, China). ELISA plates were stored at −20°C and the other reagents at 4°C. The developed plates were analyzed by an automatic microplate reader (Thermo MultiskanMK3, Thermo Fisher) and the average of three readings used from the evaluation of absorbance at 450 nm.
Paraffin Sections of Embryo sac Analysis
To analyze the embryo sac development, several flowers were selected just before pollination (florets were sampled just before pollination). The ovaries were removed and immediately fixed in cold GA-PFA solution which contained 2.5% glutaraldehyde, 2% paraformaldehyde and 50 mM PIPES (pH 7.2), at 4°C overnight as described previously (Sambrook et al., 1989). The ovaries were removed, dehydrated in a graded ethanol series (30, 50, 70, 80, 90, 95, and 100% [v/v]), with 10 min for each gradient and then embedded in paraffin. Semi thin sections (7 μm) of the embedded organ were cut with a microtome and stained with toluidine blue O. The embryo sac size was measured by Leica microscope software.
Laser Scanning Confocal Microscopy Assay
Whole-mount eosin B staining was performed using laser scanning confocal microscopy to determine embryo sac development (Zhang et al., 2003). The sample fixation method employed is described in the paraffin sectioning above. The ovaries were dissected in 70% ethanol and rehydrated sequentially in 50% ethanol, 30% ethanol and distilled water. Subsequently, the samples were pretreated in 2% KAl(SO4)2·12H2O for 20 min and then stained with 10 mg/l eosin B (C20H6N2O9Br2Na2) in 4% sucrose solution for 10–12 h at room temperature. The samples were post-treated in 2% KAl(SO4)2·12H2O for 20 min, rinsed with distilled water three times and afterwards dehydrated with a series of ethanol solutions: 30, 50, 70, 90, and 100% (v/v). The dehydrated samples were treated in a mixture of absolute ethanol and methyl salicylate (1:1 [v/v]) for 1 h, and then cleared in 100% methyl salicylate solution. The cleared samples were scanned with a Leica laser scanning confocal microscope. Excitation wavelength was 543 nm, and emission light was detected between 550 and 630 nm.
GUS Staining Assay
The GUS assay was conducted according to Lee et al. (2013). Tissue samples from pOsEMSA1::GUS transgenic plants were immersed in cold 90% acetone at −20°C for 20 min, then rinsed three times with rinse solution: 0.1 M K3Fe(CN)6, 0.1 M K4Fe(CN)6, and 0.5 M NaPO4, pH 7.2. Samples were soaked in GUS staining solution (10% Triton X-100, 20 mM X-Gluc, 0.1 M K3Fe(CN)6, 0.1 M K4Fe(CN)6 and 0.5 M NaPO4, pH 7.2) and incubated at 37°C overnight. After staining, the samples were bleached with 75% ethanol and observed under a dissecting microscope.
Gene Expression Analysis by RT-PCR
Extraction of total RNA from plant tissues at different developmental stages was performed using the TRNzol reagent (TRNzol, TianGen Biotech Co. Ltd., Beijing, China). The cDNAs were synthesized from 2 mg of total RNA according to the manufacturer's protocol (RevertAid First Strand cDNA Synthesis Kit, Thermo Fisher). Rice β-ACTIN was amplified and used as an internal standard to normalize the expression of tested genes. Six pairs of primers were used for RT-PCR (Supplementary Table 2).
Phenotype Characterization
Pollen fertility was calculated by determining the percentage of normal pollen grains against total pollen grains per spikelet. Seed fertility was the number of filled grains divided by the total number of grains per panicle evaluated for each panicle on five plants. Panicle length was measured as the average value in centimeters, from the panicle neck to the panicle tip based on an evaluation of three panicles from 10 random plants. Mature rice seeds were air-dried and stored at room temperature. Fully filled grains were used for grain length, width and weight measurement. Ten randomly chosen grains from each plant were lined up length-wise along a Vernier caliper to measure grain length, and then arranged by breadth to measure grain width. Grain weight was calculated based on 100 grains and converted to thousand-grain weight.
Results
OsEMSA1 Cloning and Characterization
The OsEMSA1 gene was cloned to allow exploring its function. We isolated the 2.6 kb putative promoter region of OsEMSA1 and the 952-bp full-length cDNA (GenBank accession number: KX503265) with 327 bp open reading frame (ORF), which encodes a protein with 108 amino acid residues (Figure 1Aa). The protein domain identification tool, SMART, identified an N-terminal signal peptide at position 1–37 and a LysM domain at the C-terminal at position 58–101 (Figures 1A,B). A search on the Protein BLAST NCBI database identified a number of homologs with a conserved LysM domain. Phylogenetic analysis showed that the protein is distributed in monocotyledons and dicotyledons but with no specific function assigned (Figure 1C). To investigate potential regulatory cis-acting elements, we analyzed the promoter region of OsEMSA1 using PlantCARE—this detailed analysis revealed that it contained 24 different cis-regulatory elements involved in various processes. In addition to the typical TATA-box and CAAT-box, there were hormone responsive elements, growth regulators, metabolism regulators and several stress-responsive regulatory elements found (Supplementary Table 1). Most of these predicted elements are involved in growth and stress responses, suggesting that the OsEMSA1 promoter may play multi-functional roles.
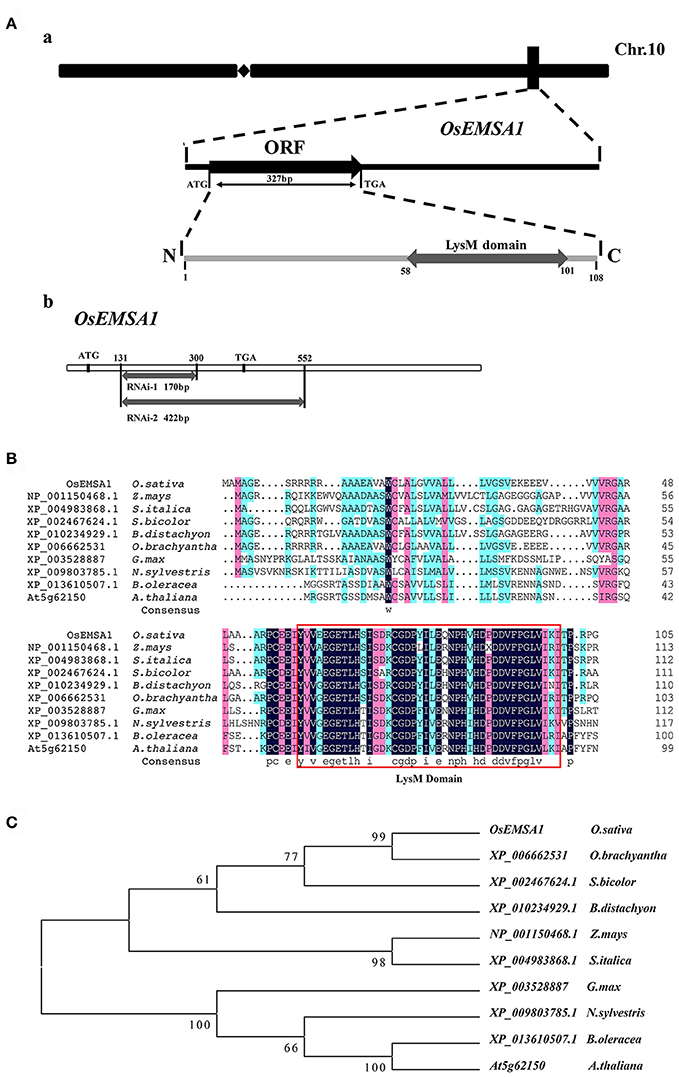
Figure 1. Molecular cloning and phylogenetic analysis. (A) Diagram of the OsEMSA1. (a) OsEMSA1 was located on chromosome 10. ORF is shown as a black box and the LysM domain as a gray box. (b) Two different fragments for RNAi construction are shown as gray boxes. (B) Sequence alignment of OsEMSA1 protein and its homologs in plants, generated with DNAMAN. The LysM domain is outlined with the red box. (C) Phylogenetic analysis of OsEMSA1 protein and its homologs in plants based on the alignment above. The phylogenetic tree was constructed by neighbor-joining method. Bootstrap value = 1,000.
Expression Pattern of OsEMSA1
Semi quantitative RT-PCR analyses using total RNA samples from various tissues at different developmental stages were performed to determine the OsEMSA1 expression profile. Different levels of transcription was detected in various organs from early vegetative to reproductive stages. Expression of OsEMSA1 was detected before pollination to 15 days after pollination. High expression was observed in roots during all developmental stages. Expression of the gene was also observed in the panicle, stem, leaf sheath, leaf blade and at the panicle initiation stage (Figure 2B). To better understand the OsEMSA1 expression pattern, the OsEMSA1 promoter was fused to the GUS reporter gene and introduced into wild type rice Ilmibyeo by Agrobacterium-mediated transformation approach. Histochemical staining of GUS showed high expression of OsEMSA1 in various tissues at different developmental stages. In consonance with RT-PCR results in roots, there was very high activity of OsEMSA1 in roots at all developmental stages. GUS activity in roots was found in the root elongation zone, lateral roots and vascular bundle (Figures 2Aa–c). GUS activity was also observed in the commissural vein of leaves and spikelet after heading stage (Figures 2Ad,g). Notably, in female reproductive organs, GUS activity was high at the early stages of flowering. Results showed that OsEMSA1 was mainly expressed in the ovary (Figure 2Ah).
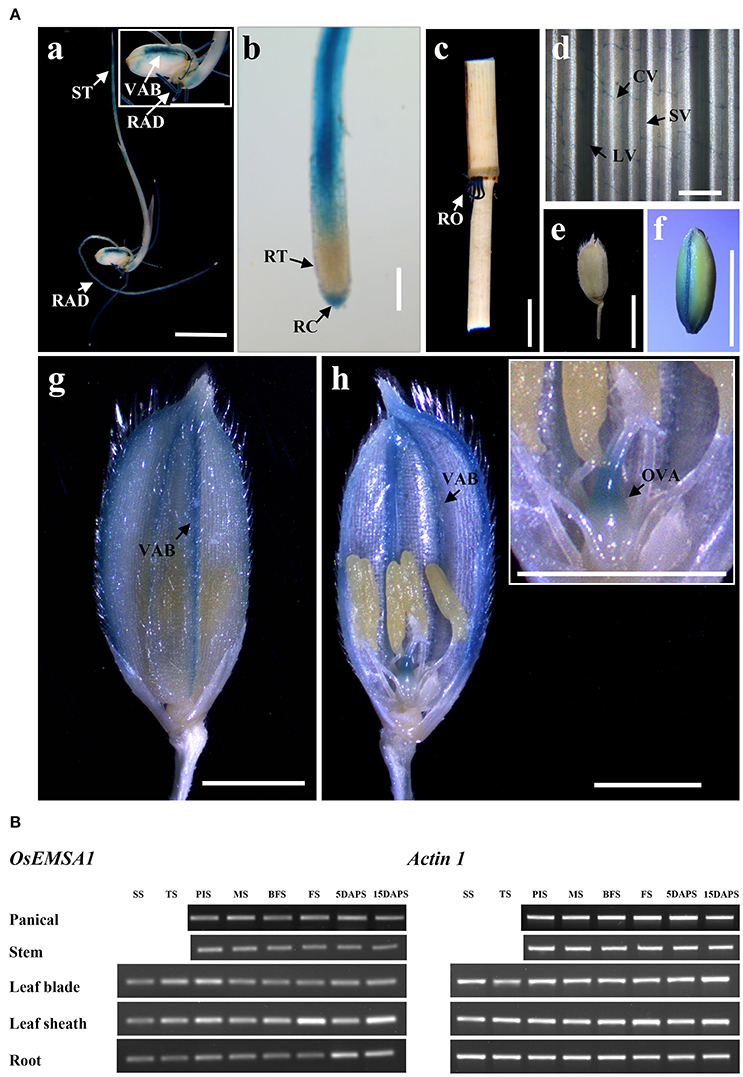
Figure 2. Expression pattern of OsEMSA1. (A) OsEMSA1 expression pattern in different tissues of pOsEMSA1::GUS transgenic rice. (a,f) GUS expression in the shoot tip, radicle and dorsal vascular bundle. (b) Elongation zone of radicle and root cap. (c) Mature root. (d) Commissural vein in leaf. (e) Mature spikelet. (g,h) Ovary and vascular bundles of glume. RAD, radicle; ST, shoot tip; VAB, vascular bundle; RT, root tip; RC, root cap; RO, root; CV, commissural vein; LV, large vein; SV, small vein; OVA, ovary. Bars = 5 mm in (a,c,e,f); 2 mm in (g,h). (B) RT-PCR analysis of OsEMSA1. SS, seedling stage; TS, tillering stage; PIS, panicle initiation stage; MS, meiotic stage; BFS, before flowering stage; FS, flowering stage; 5DAPS, 5 days after pollination stage; 15DAPS, 15 days after pollination stage.
OsEMSA1 Does Not Affect Vegetative Growth and Pollen Development
To determine whether OsEMSA1 regulates growth or reproductive processes, we generated two types of OsEMSA1 RNAi (pCaMV35S×2::OsEMSA1-RNAi1 and pCaMV35S×2::OsEMSA1-RNAi2) transgenic plants (Figure 1Ab). A significant number of RNAi transgene events were associated with completely failed sexual reproduction, and thus, could not be characterized in detail. Only those transgenic plants where reproduction succeeded at some level were selected for further characterization. The transgenic plants exhibited normal vegetative growth patterns in terms of germination, tillering and elongation. However, the overall seed-setting rate of both types of the RNAi lines was significantly decreased as compared to the wild-type (WT) (Figure 3B). Among these transgenic plants, lines R2, R4, R5, R10, R12, and R13 whose seed setting were significantly arrested, were selected as representatives for further analysis. The expression of OsEMSA1 was significantly down-regulated in these RNAi lines (Figure 3D). Panicle lengths of these transgenic lines were examined and were found to be 4 cm shorter (18.23 ± 0.66 to 22.85 ± 0.24 cm) than the WT (Figures 3A,C).
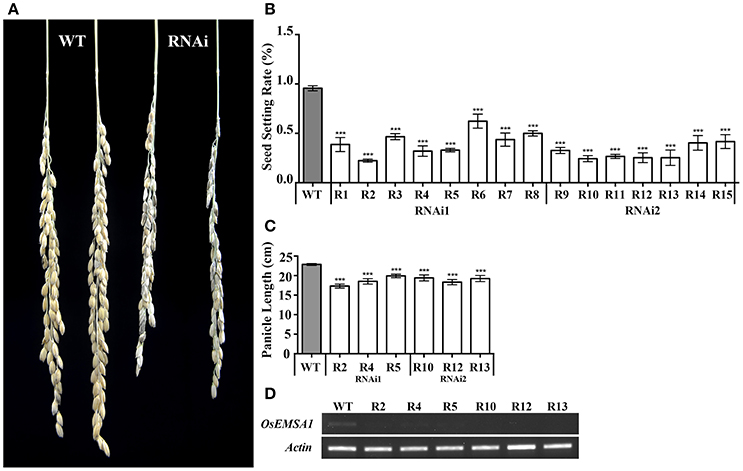
Figure 3. Biological trait comparisons of WT and OsEMSA1 RNAi lines. (A) Panicles of WT and OsEMSA1 RNAi plants. R1 to R15 are all OsEMSA1 RNAi lines. (B) Seed setting rate of T1 OsEMSA1 RNAi lines. (C) Panicle length of candidate OsEMSA1 RNAi lines. (D) OsEMSA1 expression analysis in RNAi lines. The whole-plant RNA samples were used in RT-PCR. Values are mean ± SD. Asterisks indicated significant differences (*P < 0.05) and extremely significant differences (**P < 0.01 and ***P < 0.001).
The overall decrease in seed setting rate indicated an ineffective fertilization. Effective fertilization requires functional male and female gametophytes that produce functional sex gametes. To explore the role of OsEMSA1 in male gametogenesis, we examined the development of male reproductive organs. Pollen from the representative RNAi transgenic lines and WT were examined by iodine-potassium iodide (I2-KI) and simplified Alexander's stain methods to determine fertility and viability, respectively (Figures 4Aa,b; Figures 4Ae,f). Statistical analysis showed that viability rate of pollens in RNAi lines were not different from that of WT (Figure 4B). We further determined the pollen germination and pollen tube growth in the transgenic lines (Figures 4Ac,d; Figures 4Ag,h). Consistent to WT, approximately 80–85% pollen from the transgenic lines germinated and produced pollen tubes with no morphological difference to the WT (Figure 4C).
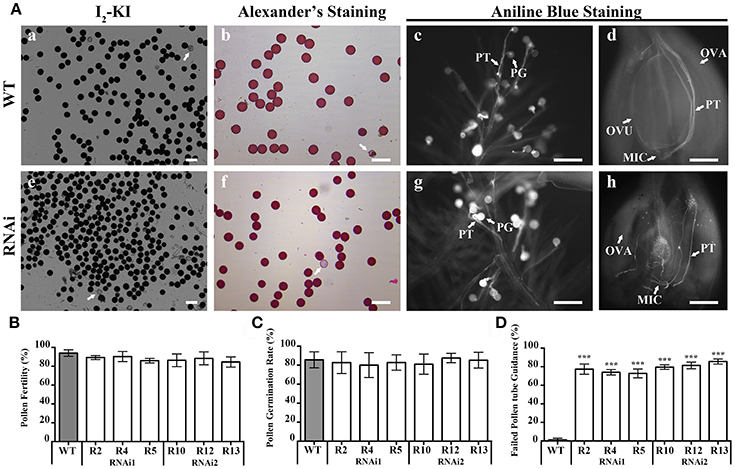
Figure 4. Pollen viability and pollen germination in WT and OsEMSA1 RNAi lines. (A) Pollen viability assay by I2-KI and Alexander's staining. Pollen germination assay by Aniline blue staining. (a–d) WT pollen grain and ovary. (e–h) RNAi plant pollen grain and ovary. Arrows indicate aborted pollen grain in (a,b,e,f). WT, wild-type; PG, pollen grain; PT, pollen tube; OVA, ovary; OVU, ovule; MIC, micropyle. Bars = 100 μm. (B) Percentage of viable pollen grain in WT plants and RNAi plants. (C) Pollen germination rate in WT plants and RNAi plants. (D) Percentage of random path pollen tube in WT plants and RNAi plants. Values are mean ± SD. Asterisks indicated extremely significant differences (***P < 0.001).
OsEMSA1 Regulates Female Gametophyte Development
Interestingly, Pollen tubes germinated and grew normally in RNAi plants. Pollen tubes entered the pistil through the stigmatic cells and reached the ovules similarly to that of the WT, indicating normal pollen tube growth and sporophytic guidance. However, within the ovary, pollen tubes of the RNAi lines behaved differently. Pollen tubes of the RNAi lines grew toward the ovule but instead of entering the micropylar opening, 80% of them seemed to lose their path and coiled randomly (Figures 4Ah,D). In some cases, pollen tubes were found to wrap around the ovules. The pollen tubes reached the micropylar end but could not enter the embryo sac, indicating that pollen tube guidance was not normal and that the RNAi lines failed to attract pollen tubes. However, pollen tubes of the WT successfully entered the female gametophyte (Figure 4Ad). Together, these results indicated that the mutation in OsEMSA1 did not affect the development and function of the male gametophyte.
To understand the reason behind the poor seed setting rate, we next examined female gametophyte development in the RNAi transgenic plants. Pistils just before pollination stage were collected from the representative RNAi and WT plants. Embryo sac development in the WT proceeded normally, showing a clear differentiation of antipodal, central cells and an egg apparatus (Figures 5Aa,d). Longitudinal sections of embryo sacs of RNAi plants showed that the gene silencing affected normal development of the embryo sac (Figures 5Ab,e; Figures 5Ac,f; Supplementary Figures 1, 2). RNAi ovary paraffin sections showed various levels of developmental and morphological defects in the embryo sac. Quantitative analysis showed that 80% of the RNAi transgenic plants harbored distorted, irregular and shrunken embryo sacs (Figure 5C). The embryo sacs of RNAi lines were degenerated and with poor differentiation of any gametophytic cells. Observations of the development of embryo sacs in RNAi transgenic plants using confocal microscopy showed that the female gametophyte failed to undergo normal nuclear divisions and cellularization during megagametogenesis (Figure 5B). These results indicated that OsEMSA1 was involved in embryo sac development in rice.
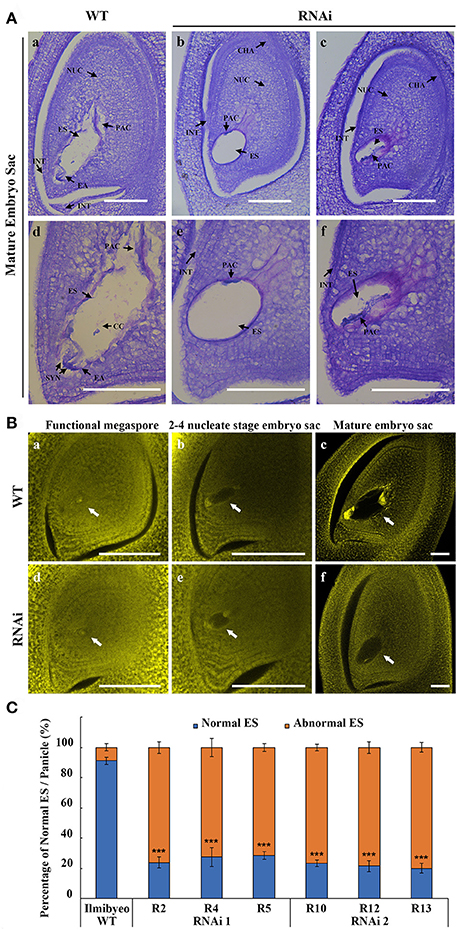
Figure 5. OsEMSA1 control of embryo sac development. (A) Longitude section of embryo sac before flowering stage. (a,d) Normal embryo sac in WT plant. (b,e and c,f) Different types of undeveloped embryo sacs in OsEMSA1 RNAi plants. (d–f) are enlarged figures of (a–c), respectively. NUC, nucellus; ES, embryo sac; PAC, possible antipodal cell; INT, integument; EA, egg apparatus; CHA, chalaza; SYN, synergid; CC, central cell; Bars = 100 μm. (B) Morphology of embryo sac during megagametogenesis phases using confocal microscopy. White arrows respectively indicate (a,d) functional megaspores, (b,e) 2–4 nucleate stage embryo sacs and (c,f) mature embryo sacs in WT and RNAi mutant plants. Bars = 100 μm. (C) Percentage of normal embryo sac in WT plants and RNAi plants. Values are mean ± SD. Asterisks indicated extremely significant differences (***P < 0.001).
OsEMSA1 Affects Hormones Level in Multiple Plant Tissues
In our previous study, fst mutation decreased the expression of several genes related to developmental and hormonal pathways. OsEMSA1 is one of the down-regulated genes in the FST network. To determine whether this gene had some role in hormone regulation, we explored the hormone contents in different tissues of the transgenic plants. Results indicated that the expression profile of OsEMSA1 affected the endogenous hormone content in different tissues of the RNAi plants at the heading stage. Gibberellic acid (GA3) and indole-3-acetic acid (IAA) content were significantly lower in leaf blade, leaf sheath, roots, and panicle for RNAi lines compared with WT (Figure 6A). We further investigated whether OsEMSA1 affected expression of genes related to hormone regulation. Four genes which are classified as hormone regulatory genes by gene annotation and which are co-down regulated along with OsEMSA1, were selected from our previous microarray data (Lee et al., 2013). OsHox24, Os.51835, OsNAC5 and Os.43929 transcript levels were examined in heading stage panicles of OsEMSA1 RNAi transgenic lines and WT by semi-quantitative RT-PCR (Kikuchi et al., 2000; Yu et al., 2005; Kawahara et al., 2013). The expression level of Os.43929 was increased, while expression level of the other three genes decreased in the mutant compared with WT (Figure 6B). These results suggested that OsEMSA1 might have some putative role in hormone regulation in rice.
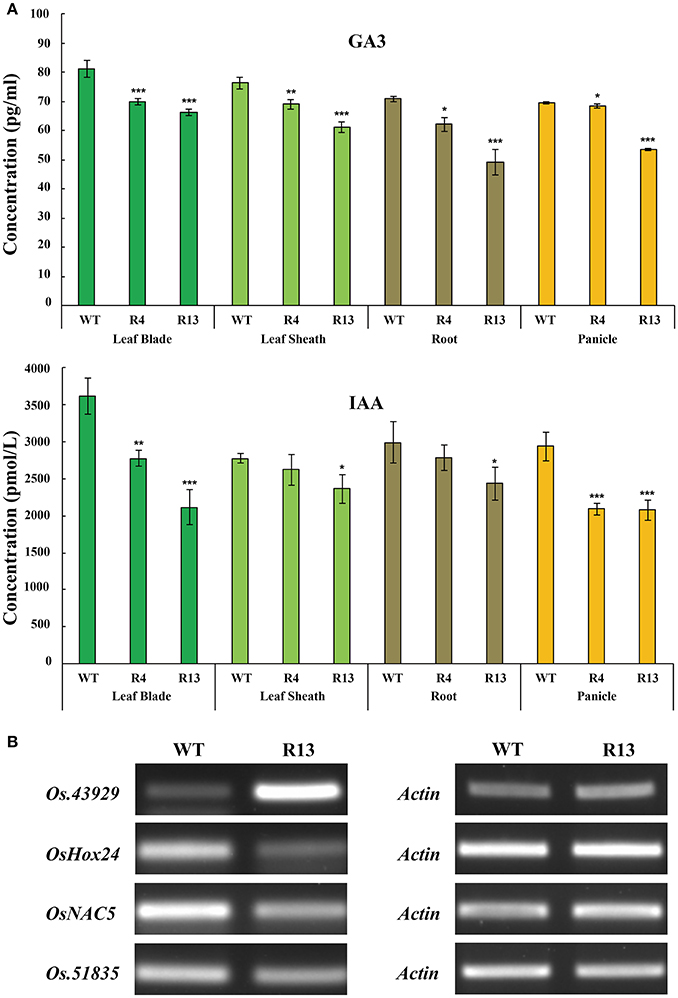
Figure 6. OsEMSA1 control of hormone distribution and expression of hormone regulator genes. (A) Hormone content comparison between WT and OsEMSA1 RNAi plants at heading stage. Content of GA3 and IAA in leaf blade, leaf sheath, root and panicle. Values are mean ± SD. Asterisks indicated significant differences (*P < 0.05) and extremely significant differences (**P < 0.01 and ***P < 0.001). (B) RT-PCR analysis of hormone regulator genes expression pattern in OsEMSA1 RNAi plants.
Discussion
Novel Function of LysM Domain-Containing Gene OsEMSA1
We showed that OsEMSA1 possessed a conserved C-terminal LysM domain and was involved in development of the embryo sac in rice. Bioinformatic analysis predicted an N-terminal signal peptide, a single LysM domain of 42 amino acid residues and encodes a protein of 108 amino acids. Sequence homology and phylogenetic analysis showed that the protein is widely distributed in both monocotyledons and dicotyledons but with no specific function assigned. Our data on the role of LysM domain-containing gene OsEMSA1 suggested that this gene may be involved in important functions not previously known. The LysM domain is a widespread protein module and has been found in plants, fungi, animals and humans (Shi et al., 2013). In plants, LysM domain-containing proteins are the second major class of highly conserved pattern recognition proteins and are involved in plant innate immunity (Gust et al., 2012; Shi et al., 2013; Tanaka et al., 2013; Wan, 2015). A typical LysM domain consists of approximately 44–65 amino acids and forms a β-α-α-β-strand secondary structure (Bateman and Bycroft, 2000). In Arabidopsis, LysM domain-containing proteins were found to be involved in peptidoglycan and chitin recognition and in turn mediated immunity against pathogen invasion (Miya et al., 2007; Willmann et al., 2011). Mutation in any of the genes significantly compromised Arabidopsis defense responses, leading to increased susceptibility to pathogens (Wan et al., 2008). Rice blast and bacterial blight are the two most devastating rice diseases, and cause significant yield losses around the globe (Liu et al., 2014). Several rice LysM domain-containing proteins directly or indirectly recognize the variable pathogen fragments and trigger defense responses. Silencing these genes considerably increases susceptibility of rice to bacterial and fungal pathogens by blockage of defense mechanisms (Kaku et al., 2006; Shimizu et al., 2010; Liu et al., 2014). In vertebrates, six zebrafish LysM domain-containing genes of two distinct sub-families called LysMD and OXR were identified and found to be strongly expressed in zebrafish embryos, but none of these genes was responsive to challenge with bacterial pathogens (Laroche et al., 2013). To date, no report on the role of LysM domain-containing proteins in plant sexual reproduction has been presented. In this study, we identified the involvement of OsEMSA1 in sexual reproduction of rice, which offers a new insight into the functions of LysM domain-containing proteins.
OsEMSA1 Silencing Disrupts Embryo sac Development in Rice
Our results on male gametogenesis indicated that the OsEMSA1 did not affect anther development. The RNAi transgenic lines produced viable pollen that germinated normally. The growth of the pollen tube toward the pistil indicated that pollen tube growth and sporophytic guidance were similar to the WT. We further investigated development of the embryo sac and observed distinct abnormalities in the embryo sac of the RNAi transgenic plants. Embryo sac development was severely arrested and was much smaller and shrunken with an irregular shape. The embryo sacs were empty and contained no obvious egg apparatus compared with the WT. The embryo sacs of the RNAi lines failed to attract the pollen tubes which after reaching the transmitting tract seemed to lose their path, started abnormal growth patterns and failed to enter the micropylar ending. Successful fertilization requires controlled growth and guidance of the pollen tube until it enters the micropylar opening of the female gametophyte (Palanivelu and Tsukamoto, 2012). Pollen tube guidance requires a complex signaling network that involves gametophytic as well as sporophytic tissues of the female gametophyte (Kasahara et al., 2005; Márton et al., 2005; Chen et al., 2007; Alandete-Saez et al., 2008; Okuda et al., 2009; González-gutiérrez et al., 2014). Mutants with a defective female gametophyte fail to fertilize and develop seeds (Palanivelu and Tsukamoto, 2012). Various studies have mentioned that the embryo sac regulates the micropylar pollen tube guidance. Ling et al. reported the role of SUMO E3 ligase (SIZ1) in the functioning of the mature embryo sac in Arabidopsis. SIZ1 mutants developed abnormal embryo sacs, which failed to attract pollen tubes and resulted in decreased seed set (Ling et al., 2012). Arabidopsis ovules carrying magatama3 failed to attract pollen tubes due to delays in embryo sac maturation, indicating that pollen tube guidance signals originated only from mature ovules (Shimizu et al., 2008). Pollen tube guidance 1 (PTB1) is responsible for the sporophytic guidance of pollen tube in rice. PTB1 encodes a RING-type E3 ubiquitin ligase and is expressed in the stigma and style. PTB1 is a domestication-related gene and is thought to have been under human selection during rice domestication because it regulates the panicle seed-setting rate (Li et al., 2013). The secreted chemotactic attractants of the female gametophyte have been identified in many plants (Shimizu and Okada, 2000; Higashiyama et al., 2001). However, no study has yet identified any gene in rice involved in gametophytic cell-cell communication. Our study preclude distinguishing between sporophytic vs. gametophytic activity for the RNAi constructs however, the OsEMSA1 RNAi transgenic plants provide a great opportunity for studying the precise origin of male-female chemotactic signals and the underlying mechanisms involved in sexual reproduction in rice.
To determine whether OsEMSA1 has some role in hormone regulation in rice, we examined the endogenous content of GA3 and IAA in different tissues of RNAi transgenic plants. Results showed that OsEMSA1 affected the hormone content in different tissues. We further investigated whether OsEMSA1 had some effect on other genes related to hormone regulation. We selected four genes that were also down regulated in our previous fst microarray data (Lee et al., 2013). Gene annotation of our microarray data suggested them to be involved in hormone regulation. Among the four selected genes, the gene OsHox24 and OsNAC5 are previously identified to be hormone responsive genes (Sperotto et al., 2009; Bhattacharjee et al., 2016, 2017). OsHox24 is a member of homeobox transcription factor family and play important role in rice plant growth and development (Bhattacharjee et al., 2017). Previous studies have identified the role of OsHox24 in abiotic stress responses in rice by regulating the expression of other stress responsive genes (Bhattacharjee et al., 2016, 2017). OsHox24 has been identified as a hormone responsive gene and is involved in ABA, GA, SA, or IAA-signaling pathway (Olsson et al., 2004; Bhattacharjee et al., 2016). OsNAC5 is a member of the NAC family transcription factors that regulates abiotic stress responses in rice by modulating the expression of stress-responsive genes (Sperotto et al., 2009; Takasaki et al., 2010; Song et al., 2011). Previous study has identified an ABA- dependent expression of OsNAC5 during grain filling stage in rice (Sperotto et al., 2009). Our independent experiments on the two co-downregulated genes, Os.51835 and Os.43929, indicated their involvement in hormone regulation in rice (Supplementary Figure 3). Our results indicated that suppression of Os.51835 and Os.43929 expression resulted in a reduction of GA3 and IAA content in transgenic plants. To investigate the relation of OsEMSA1 with the other co-downregulated genes, we performed RT-PCR analysis. Our semi-quantitative RT-PCR analysis of RNAi mutant panicles at heading stage showed that OsEMSA1 also regulated the expression of genes involved in hormone regulation and suggests that OsEMSA1 was involved in hormone regulation in rice. Phytohormones control different developmental processes in plants. Auxin play important role in ovule patterning in the female gametophyte of Arabidopsis (Pagnussat et al., 2009). It was observed that syncitial embryo sac cell fate can be regulated by asymmetrical sporophytic and gametophytic auxin gradient (Pagnussat et al., 2009; Sundaresan and Alandete-Saez, 2010; Panoli et al., 2015). Previous studies showed that auxin source (IAA) is located in the sporophytic tissue at early stage embryo sac then transfer to micropylar end of female gametophyte. The egg apparatus fate would correspond to the highest auxin concentration which is formed in the syncytial embryo sac at the micropylar pole and antipodal fate to the lowest at the chalazal pole (Pagnussat et al., 2007, 2009; Sundaresan and Alandete-Saez, 2010). Although numerous studies have highlighted the role of phytohormones in male and female reproductive organogenesis, but these studies on the embryo sac development in monocots in general and in rice in particular are not comprehensive. Much work needs to be done to explore the molecular mechanisms involved in phytohormones mediated organogenesis. Our present finding supports our previous studies on fst mutant in rice whereby the mutant developed defective ovules and complete abortion of the embryo. The precise molecular network of FST and its associated genes controlling the various developmental processes in rice is yet to be revealed.
Our results revealed that OsEMSA1 had a definite role in the development and cellularization of female gametophytic cells in rice. Gene silencing resulted in a defective and degenerated embryo sac that failed to attract pollen tubes. Our results demonstrated that OsEMSA1 was directly or indirectly play some role in the endogenous hormone regulation and embryo sac development in rice. Our study provides a novel function of LysM domain-containing proteins in female gametophyte development of rice.
Author Contributions
DL and LC conceived the original project and research plans; DL, LC, and CL supervised the experiments; QZ and XZ designed the experiments and performed most of the experiments; QZ, XZ, and SN analyzed the data; SN, WD, XG, and HZ provided technical assistance. SN and QZ wrote the article with contributions of all the authors; All authors supervised and complemented the writing. All authors agree to be accountable for the content of the work.
Funding
1. National Key Research and Development Program of China (Grant Number: 2016YFD0101101-5).
2. National Natural Science Foundation of China (Grant Number: U1136604).
3. New Academic Researcher Award for Doctoral Candidates of Yunnan Province (Grant Number: A3007962).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to J. Yang for technical assistant of laser scanning confocal microscopy and Henry Akrofi Doku for his help of manuscript revision. We thank X. L. Tan and L. Z. Xiong for scientific discussions and insightful suggestions. We thank J. C. Wen, J. Shi, H. Y. Xu, Y. L. Tan, J. Li, M. H. Sohn, H. B. Xiong, Z. F. Wei, Y. G. Lv, T. F. Wu, L. D. Zhang, W. Li, C. Wu, X. M. Yin, C. M. Hu, F. Luo, Y. X. Fu, K. B. Wang, and S. Q. Liu for their technical support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01596/full#supplementary-material
References
Akcapinar, G. B., Kappel, L., Sezerman, O. U., and Seiboth, V. S. (2015). Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr. Genet. 61, 103–113. doi: 10.1007/s00294-014-0471-9
Alandete-Saez, M., Ron, M., and McCormick, S. (2008). GEX3, expressed in the male gametophyte and in the egg cell of Arabidopsis thaliana, is essential for micropylar pollen tube guidance and plays a role during early embryogenesis. Mol. Plant 1, 586–598. doi: 10.1093/mp/ssn015
Bateman, A., and Bycroft, M. (2000). The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD)1. J. Mol. Biol. 299, 1113–1119. doi: 10.1006/jmbi.2000.3778
Bencivenga, S., Simonini, S., Benkova, E., and Colombo, L. (2012). The transcription factors bel1 and spl are required for cytokinin and auxin signaling during ovule development in Arabidopsis. Plant Cell 24, 2886–2897. doi: 10.1105/tpc.112.100164
Bhattacharjee, A., Khurana, J. P., and Jain, M. (2016). Characterization of rice homeobox genes OsHox22 and OsHox24, and over-expression of OsHox24 in transgenic Arabidopsis suggest their role in abiotic stress response. Front. Plant Sci. 7:627. doi: 10.3389/fpls.2016.00627
Bhattacharjee, A., Sharma, R., and Jain, M. (2017). Over-Expression of OsHOX24 confers enhanced susceptibility to abiotic stresses in transgenic rice via modulating stress-responsive gene expression. Front. Plant Sci. 8:628. doi: 10.3389/fpls.2017.00628
Buist, G., Steen, A., Kok, J., and Kuipers, O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido) glycans. Mol. Microbiol. 68, 838–847. doi: 10.1111/j.1365-2958.2008.06211.x
Chen, Y. H., Li, H. J., Shi, D. Q., Yuan, L., Liu, J., Sreenivasan, R., et al. (2007). The central cell plays a critical role in pollen tube guidance in Arabidopsis. Plant Cell Online 19, 3563–3577. doi: 10.1105/tpc.107.053967
Cheng, C. Y., Mathews, D. E., Schaller, G. E., and Kieber, J. J. (2013). Cytokinin-dependent specification of the functional megaspore in the Arabidopsis female gametophyte. Plant J. 73, 929–940. doi: 10.1111/tpj.12084
Cheung, A. Y., Wang, H., and Wu, H. M. (1995). A floral transmitting tissue-specific glycoprotein attracts pollen tubes and stimulates their growth. Cell 82, 383–393. doi: 10.1016/0092-8674(95)90427-1
Deng, Y., Dong, H., Mu, J., Ren, B., Zheng, B., Ji, Z., et al. (2010). Arabidopsis histidine kinase CKI1 acts upstream of histidine phosphotransfer proteins to regulate female gametophyte development and vegetative growth. Plant Cell 22, 1232–1248. doi: 10.1105/tpc.108.065128
Drews, G. N., Lee, D., and Christensen, C. A. (1998). Genetic analysis of female gametophyte development and function. Plant Cell 10, 5–17. doi: 10.1105/tpc.10.1.5
Fiebig, A., Mayfield, J. A., Miley, N. L., Chau, S., Fischer, R. L., and Preuss, D. (2000). Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12, 2001–2008. doi: 10.1105/tpc.12.10.2001
González-gutiérrez, A. G., Gutiérrez-mora, A., and Rodríguez-garay, B. (2014). Embryo sac formation and early embryo development in Agave tequilana (Asparagaceae). Springerplus 3, 1–11. doi: 10.1186/2193-1801-3-575
Gust, A. A., Willmann, R., Desaki, Y., Grabherr, H. M., and Nürnberger, T. (2012). Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502. doi: 10.1016/j.tplants.2012.04.003
Higashiyama, T., Kuroiwa, H., Kawano, S., and Kuroiwaa, T. (1998). Guidance in vitro of the pollen tube to the naked embryo sac of Torenia fournieri. Plant Cell 10, 2019–2031. doi: 10.1105/tpc.10.12.2019
Higashiyama, T., Yabe, S., Sasaki, N., Nishimura, Y., Miyagishima, S., Kuroiwa, H., et al. (2001). Pollen tube attraction by the synergid cell. Science 293, 1480–1483. doi: 10.1126/science.1062429
Hirano, K., Aya, K., Hobo, T., Sakakibara, H., Kojima, M., Shim, R. A., et al. (2008). Comprehensive transcriptome analysis of phytohormone biosynthesis and signaling genes in microspore/pollen and tapetum of rice. Plant Cell Physiol. 49, 1429–1450. doi: 10.1093/pcp/pcn123
Hulskamp, M., Schneitz, K., and Pruitt, R. (1995). Genetic evidence for a long-range activity that directs pollen tube guidance in Arabidopsis. Plant Cell 7, 57–64. doi: 10.1105/tpc.7.1.57
Kaku, H., Nishizawa, Y., Ishii-Minami, N., Akimoto-Tomiyama, C., Dohmae, N., Takio, K., et al. (2006). Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. U.S.A. 103, 11086–11091. doi: 10.1073/pnas.0508882103
Kasahara, R. D., Portereiko, M. F., Sandaklie-Nikolova, L., Rabiger, D. S., and Drews, G. N. (2005). MYB98 is required for pollen tube guidance and synergid cell differentiation in Arabidopsis. Plant Cell 17, 2981–2992. doi: 10.1105/tpc.105.034603
Kawahara, Y., de la Bastide, M., Hamilton, J. P., Kanamori, H., McCombie, W. R., Ouyang, S., et al. (2013). Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 6:4. doi: 10.1186/1939-8433-6-4
Kikuchi, K., Ueguchi-Tanaka, M., Yoshida, K. T., Nagato, Y., Matsusoka, M., and Hirano, H. Y. (2000). Molecular analysis of the NAC gene family in rice. Mol. Gen. Genet. 262, 1047–1051. doi: 10.1007/PL00008647
Kinoshita-Tsujimura, K., and Kakimoto, T. (2011). Cytokinin receptors in sporophytes are essential for male and female functions in Arabidopsis thaliana. Plant Signal. Behav. 6, 66–71. doi: 10.4161/psb.6.1.13999
Laroche, F. J. F., Tulotta, C., Lamers, G. E. M., Meijer, A. H., Yang, P., Verbeek, F. J., et al. (2013). The embryonic expression patterns of zebrafish genes encoding LysM-domains. Gene Expr. Patterns 13, 212–224. doi: 10.1016/j.gep.2013.02.007
Lee, D. S., Chen, L. J., Li, C. Y., Liu, Y., Tan, X. L., Lu, B. R., et al. (2013). The Bsister MADS gene FST determines ovule patterning and development of the zygotic embryo and endosperm. PLoS ONE 8:e58748. doi: 10.1371/journal.pone.0058748
Li, M., Xu, W., Yang, W., Kong, Z., and Xue, Y. (2007). Genome-wide gene expression profiling reveals conserved and novel molecular functions of the stigma in rice. Plant Physiol. 144, 1797–1812. doi: 10.1104/pp.107.101600
Li, S., Li, W., Huang, B., Cao, X., Zhou, X., Ye, S., et al. (2013). Natural variation in PTB1 regulates rice seed setting rate by controlling pollen tube growth. Nat. Commun. 4:2793. doi: 10.1038/ncomms3793
Ling, Y., Zhang, C., Chen, T., Hao, H., Liu, P., Bressan, R. A., et al. (2012). Mutation in SUMO E3 ligase, SIZ1, disrupts the mature female gametophyte in Arabidopsis. PLoS ONE 7:e29470. doi: 10.1371/journal.pone.0029470
Liu, B., Li, J. F., Ao, Y., Qu, J., Li, Z., Su, J., et al. (2012). Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell 24, 3406–3419. doi: 10.1105/tpc.112.102475
Liu, W., Liu, J., Triplett, L., Leach, J. E., and Wang, G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 52, 213–241. doi: 10.1146/annurev-phyto-102313-045926
Márton, M. L., Cordts, S., Broadhvest, J., and Dresselhaus, T. (2005). Micropylar pollen tube guidance by egg apparatus 1 of maize. Science 307, 573–576. doi: 10.1126/science.1104954
Miya, A., Albert, P., Shinya, T., Desaki, Y., Ichimura, K., Shirasu, K., et al. (2007). CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 104, 19613–19618. doi: 10.1073/pnas.0705147104
Mollet, J. C., Park, S. Y., Nothnagel, E. A., and Lord, E. M. (2000). A lily stylar pectin is necessary for pollen tube adhesion to an in vitro stylar matrix. Plant Cell 12, 1737–1750. doi: 10.1105/tpc.12.9.1737
Mori, T., Kuroiwa, H., Higashiyama, T., and Kuroiwa, T. (2006). GENERATIVE CELL SPECIFIC 1 is essential for angiosperm fertilization. Nat. Cell Biol. 8, 64–71. doi: 10.1038/ncb1345
Olsson, A. S. B., Engström, P., and Söderman, E. (2004). The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol. Biol. 55, 663–677. doi: 10.1007/s11103-004-1581-4
Okuda, S., Tsutsui, H., Shiina, K., Sprunck, S., Takeuchi, H., Yui, R., et al. (2009). Defensin-like polypeptide LUREs are pollen tube attractants secreted from synergid cells. Nature 458, 357–361. doi: 10.1038/nature07882
Pagnussat, G. C., Alandete-Saez, M., Bowman, J. L., and Sundaresan, V. (2009). Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324, 1684–1689. doi: 10.1126/science.1167324
Pagnussat, G. C., Yu, H. J., Ngo, Q. A., Rajani, S., Mayalagu, S., Johnson, C. S., et al. (2005). Genetic and molecular identification of genes required for female gametophyte development and function in Arabidopsis. Development 132, 603–614. doi: 10.1242/dev.01595
Pagnussat, G. C., Yu, H. J., and Sundaresan, V. (2007). Cell-fate switch of synergid to egg cell in Arabidopsis eostre mutant embryo sacs arises from misexpression of the BEL1-like homeodomain gene BLH1. Plant Cell 19, 3578–3592. doi: 10.1105/tpc.107.054890
Palanivelu, R., Brass, L., Edlund, A. F., and Preuss, D. (2003). Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114, 47–59. doi: 10.1016/S0092-8674(03)00479-3
Palanivelu, R., and Tsukamoto, T. (2012). Pathfinding in angiosperm reproduction: pollen tube guidance by pistils ensures successful double fertilization. Wiley Interdiscip. Rev. Dev. Biol. 1, 96–113. doi: 10.1002/wdev.6
Panoli, A., Martin, M. V., Alandete-Saez, M., Simon, M., Neff, C., Swarup, R., et al. (2015). Auxin import and local auxin biosynthesis are required for mitotic divisions, cell expansion and cell specification during female gametophyte development in Arabidopsis thaliana. PLoS ONE 10:e0126164. doi: 10.1371/journal.pone.0126164
Park, J. I., Hakozaki, H., Endo, M., Takada, Y., Ito, H., Uchida, M., et al. (2006). Molecular characterization of mature pollen-specific genes encoding novel small cysteine-rich proteins in rice (Oryza sativa L.). Plant Cell Rep. 25, 466–474. doi: 10.1007/s00299-005-0077-2
Peterson, R., Slovin, J. P., and Chen, C. (2010). A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 1, 66–69. doi: 10.4081/pb.2010.e13
Ray, S. M., Park, S. S., and Ray, A. (1997). Pollen tube guidance by the female gametophyte. Development 124, 2489–2498.
Reiser, L., and Fischer, R. (1993). The ovule and the embryo sac. Plant Cell 5, 1291–1301. doi: 10.1105/tpc.5.10.1291
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. doi: 10.1128/AEM.68.3.1232
Schaller, G. E., Bishopp, A., and Kieber, J. J. (2015). The yin-yang of hormones: cytokinin and auxin interactions in plant development. Plant Cell 27, 44–63. doi: 10.1105/tpc.114.133595
Shi, X. Z., Zhou, J., Lan, J. F., Jia, Y. P., Zhao, X. F., and Wang, J. X. (2013). A Lysin motif (LysM)-containing protein functions in antibacterial responses of red swamp crayfish, Procambarus clarkii. Dev. Comp. Immunol. 40, 311–319. doi: 10.1016/j.dci.2013.03.011
Shimizu, K. K., Ito, T., Ishiguro, S., and Okada, K. (2008). MAA3 (MAGATAMA3) helicase gene is required for female gametophyte development and pollen tube guidance in Arabidopsis thaliana. Plant Cell Physiol. 49, 1478–1483. doi: 10.1093/pcp/pcn130
Shimizu, K. K., and Okada, K. (2000). Attractive and repulsive interactions between female and male gametophytes in Arabidopsis pollen tube guidance. Development 127, 4511–4518.
Shimizu, T., Nakano, T., Takamizawa, D., Desaki, Y., Ishii-Minami, N., Nishizawa, Y., et al. (2010). Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. doi: 10.1111/j.1365-313X.2010.04324.x
Song, S. Y., Chen, Y., Chen, J., Dai, X. Y., and Zhang, W. H. (2011). Physiological mechanisms underlying OsNAC5-dependent tolerance of rice plants to abiotic stress. Planta 234:331. doi: 10.1007/s00425-011-1403-2
Sperotto, R. A., Ricachenevsky, F. K., Duarte, G. L., Boff, T., Lopes, K. L., Sperb, E. R., et al. (2009). Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 230:985. doi: 10.1007/s00425-009-1000-9
Sundaresan, V., and Alandete-Saez, M. (2010). Pattern formation in miniature: the female gametophyte of flowering plants. Development 137, 179–189. doi: 10.1242/dev.030346
Takasaki, H., Maruyama, K., Kidokoro, S., Ito, Y., Fujita, Y., Shinozaki, K., et al. (2010). The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol. Genet. Genomics 284:173. doi: 10.1007/s00438-010-0557-0
Takeuchi, H., and Higashiyama, T. (2016). Tip-localized receptors control pollen tube growth and LURE sensing in Arabidopsis. Nature 531, 245–248. doi: 10.1038/nature17413
Tanaka, K., Nguyen, C. T., Liang, Y., Cao, Y., and Stacey, G. (2013). Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal. Behav. 8:e22598. doi: 10.4161/psb.22598
Wan, J. (2015). Diverse roles of Lysin-motif (LysM) proteins in mediating plant-microbe interactions. Walailak J. 12, 631–641. doi: 10.14456/WJST.2015.65
Wan, J., Zhang, X. C., Neece, D., Ramonell, K. M., Clough, S., Kim, S. Y., et al. (2008). A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20, 471–481. doi: 10.1105/tpc.107.056754
Wang, H., Wu, H., and Cheung, A. (1993). Development and pollination regulated accumulation and glycosylation of a stylar transmitting tissue-specific proline-rich protein. Plant Cell 5, 1639–1650. doi: 10.1105/tpc.5.11.1639
Wang, N., Huang, H. J., Ren, S. T., Li, J. J., Sun, Y., Sun, D. Y., et al. (2012). The rice wall-associated receptor-like kinase gene OsDEES1 plays a role in female gametophyte development. Plant Physiol. 160, 696–707. doi: 10.1104/pp.112.203943
Wang, T., Liang, L., Xue, Y., Jia, P. F., Chen, W., Zhang, M. X., et al. (2016). A receptor heteromer mediates the male perception of female attractants in plants. Nature 531, 241–244. doi: 10.1038/nature16975
Willmann, R., Lajunen, H. M., Erbs, G., Newman, M., Kolb, D., and Tsuda, K. (2011). Mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 108, 19824–19829. doi: 10.1073/pnas.1112862108
Wu, H. M., Wong, E., Ogdahl, J., and Cheung, A. Y. (2000). A pollen tube growth-promoting arabinogalactan protein from Nicotiana alata is similar to the tobacco TTS protein. Plant J. 22, 165–176. doi: 10.1046/j.1365-313x.2000.00731.x
Yamaki, S., Nagato, Y., Kurata, N., and Nonomura, K. I. (2011). Ovule is a lateral organ finally differentiated from the terminating floral meristem in rice. Dev. Biol. 351, 208–216. doi: 10.1016/j.ydbio.2010.12.006
Yoshida, K. T., Endo, M., Nakazono, M., Fukuda, H., Demura, T., Tsuchiya, T., et al. (2005). cDNA microarray analysis of gene expression changes during pollination, pollen-tube elongation, fertilization, and early embryogenesis in rice pistils. Sex. Plant Reprod. 17, 269–275. doi: 10.1007/s00497-004-0238-4
Yu, J., Wang, J., Lin, W., Li, S., Li, H., Zhou, J., et al. (2005). The genomes of Oryza sativa: a history of duplications. PLoS Biol. 3:e38. doi: 10.1371/journal.pbio.0030038
Keywords: rice (Oryza sativa L.), OsEMSA1, female gametophyte, embryo sac development, LysM domain
Citation: Zhu Q, Zhang X-L, Nadir S, DongChen W-H, Guo X-Q, Zhang H-X, Li C-Y, Chen L-J and Lee D-S (2017) A LysM Domain-Containing Gene OsEMSA1 Involved in Embryo sac Development in Rice (Oryza sativa L.). Front. Plant Sci. 8:1596. doi: 10.3389/fpls.2017.01596
Received: 15 April 2017; Accepted: 30 August 2017;
Published: 20 September 2017.
Edited by:
Michael J. Scanlon, Cornell University, United StatesReviewed by:
Sharon Ann Kessler, Purdue University, United StatesJohn E. Fowler, Oregon State University, United States
Copyright © 2017 Zhu, Zhang, Nadir, DongChen, Guo, Zhang, Li, Chen and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Juan Chen, 964136487@qq.com
Dong-Sun Lee, dong_east@ynu.edu.kr
†These authors have contributed equally to this work.
 Qian Zhu
Qian Zhu Xiao-Ling Zhang
Xiao-Ling Zhang Sadia Nadir1,2
Sadia Nadir1,2 Li-Juan Chen
Li-Juan Chen